Contents - Introduction - Prerequisites - Python module skeleton - Feature Selection - Variance threshold - Univariate feature selection - Recursive Feature Elimination (RFE) - Resources
Introduction
Previous posts in this series on machinelearning cover an introduction to machinelearning and algorithms in machinelearning; in this post you are going to look at how to discriminate between the independent features and select only the most relevant for predictive modelling. As in the earlier posts, also this post evolves around building a Python module (.py). If you want to jump straight to feature selection, I recommend the Feature Selection in Python with Scikit-Learn page by Jason Brownlee.
You can copy and paste the code in the post, but you might have to edit some parts: indentations does not always line up, quotations marks can be of different kinds and are not always correct when published/printed as html text, and the same also happens with underscores and slashes.
The complete code is also available here.
Prerequisites
The prerequisites are the same as in the previous posts in this series: a Python environment with numpy, pandas, sklearn (Scikit learn) and matplotlib installed.
Python module skeleton
We are going to start with a similar Python module skeleton as in the previous post. If you are not familiar with the Python code below, you can build the skeleton step by step by following the initial post in this series, otherwise just copy and paste the code below to an empty Python module (.py file). The skeleton is set up for using the dataset on Boston house prices introduced in the previous posts.
import numpy as np
import pandas as pd
from sklearn import model_selection
from sklearn import linear_model
from sklearn.neighbors import KNeighborsRegressor
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.svm import SVR
from sklearn.metrics import mean_squared_error, r2_score
import matplotlib
matplotlib.use('TkAgg')
from matplotlib import pyplot
class RegressionModels:
'''Machinelearning using regression models
'''
def __init__(self, columns,target):
'''creates an instance of RegressionModels
'''
self.columns = columns
self.target = target
#create an empty dictionary for features to be discarded by each model
self.modelDiscardD = {}
def ImportUrlDataset(self,url):
self.dataframe = pd.read_csv(url, delim_whitespace=True, names=self.columns)
def ExtractDf(self,omitL):
#extract the target column as y
self.y = self.dataframe[target]
#appeld the target to the list of features to be omitted
omitL.append(self.target)
#define the list of data to use
self.columnsX = [item for item in self.dataframe.columns if item not in omitL]
#extract the data columns as X
self.X = self.dataframe[self.columnsX]
def PlotRegr(self, obs, pred, title, color='black'):
pyplot.xticks(())
pyplot.yticks(())
fig, ax = pyplot.subplots()
ax.scatter(obs, pred, edgecolors=(0, 0, 0), color=color)
ax.plot([obs.min(), obs.max()], [obs.min(), obs.max()], 'k--', lw=3)
ax.set_xlabel('Observations')
ax.set_ylabel('Predictions')
pyplot.title(title)
pyplot.show()
def ModelSelectSet(self,modD):
self.models = []
if 'OLS' in modD:
self.models.append(('OLS', linear_model.LinearRegression(**modD['OLS'])))
self.modelDiscardD['OLS'] = []
if 'TheilSen' in modD:
self.models.append(('TheilSen', linear_model.TheilSenRegressor(**modD['TheilSen'])))
self.modelDiscardD['TheilSen'] = []
if 'Huber' in modD:
self.models.append(('Huber', linear_model.HuberRegressor(**modD['Huber'])))
self.modelDiscardD['Huber'] = []
if 'KnnRegr' in modD:
self.models.append(('KnnRegr', KNeighborsRegressor( **modD['KnnRegr'])))
self.modelDiscardD['KnnRegr'] = []
if 'DecTreeRegr' in modD:
self.models.append(('DecTreeRegr', DecisionTreeRegressor(**modD['DecTreeRegr'])))
self.modelDiscardD['DecTreeRegr'] = []
if 'SVR' in modD:
self.models.append(('SVR', SVR(**modD['SVR'])))
self.modelDiscardD['SVR'] = []
if 'RandForRegr' in modD:
self.models.append(('RandForRegr', RandomForestRegressor( **modD['RandForRegr'])))
self.modelDiscardD['RandForRegr'] = []
def RegrModTrainTest(self, testsize=0.3, plot=True):
#Split the data into training and test substes
X_train, X_test, y_train, y_test = model_selection.train_test_split(self.X, self.y, test_size=testsize)
#Loop over the defined models
for m in self.models:
#Retrieve the model name and the model itself
name,mod = m
#Remove the features listed in the modelDiscarD
self.ExtractDf(self.modelDiscardD[name])
#Fit the model
mod.fit(X_train, y_train)
#Predict the independent variable in the test subset
predict = mod.predict(X_test)
#Print out the model name
print 'Model: %s' %(name)
#Print out RMSE
print(" Mean squared error: %.2f" \
% mean_squared_error(y_test, predict))
#Print explained variance score: 1 is perfect prediction
print(' Variance score: %.2f' \
% r2_score(y_test, predict))
if plot:
title = ('Model: %(mod)s; RMSE: %(rmse)2f; r2: %(r2)2f' \
% {'mod':name,'rmse':mean_squared_error(y_test, predict),'r2': r2_score(y_test, predict)} )
self.PlotRegr(y_test, predict, title, color='green')
def RegrModKFold(self,folds=10, plot=True):
#set the kfold
kfold = model_selection.KFold(n_splits=folds)
for m in self.models:
#Retrieve the model name and the model itself
name,mod = m
#Remove the features listed in the modelDiscarD
self.ExtractDf(self.modelDiscardD[name])
#cross_val_predict returns an array of the same size as `y` where each entry
#is a prediction obtained by cross validation:
predict = model_selection.cross_val_predict(mod, self.X, self.y, cv=kfold)
#to retriece regressions scores, use cross_val_score
scoring = 'r2'
r2 = model_selection.cross_val_score(mod, self.X, self.y, cv=kfold, scoring=scoring)
#The correlation coefficient
#Print out the model name
print 'Model: %s' %(name)
#Print out correlation coefficients
print(' Regression coefficients: \n', r2)
#Print out RMSE
print("Mean squared error: %.2f" \
% mean_squared_error(self.y, predict))
#Explained variance score: 1 is perfect prediction
print('Variance score: %.2f' \
% r2_score(self.y, predict))
if plot:
title = ('Model: %(mod)s; RMSE: %(rmse)2f; r2: %(r2)2f' \
% {'mod':name,'rmse':mean_squared_error(self.y, predict),'r2': r2_score(self.y, predict)} )
self.PlotRegr(self.y, predict, title, color='blue')
if __name__ == ('__main__'):
columns = ['CRIM', 'ZN', 'INDUS', 'CHAS', 'NOX', 'RM', 'AGE', 'DIS', 'RAD', 'TAX', 'PTRATIO', 'B', 'LSTAT', 'MEDV']
target = 'MEDV'
regmods = RegressionModels(columns, target)
regmods.ImportUrlDataset('https://archive.ics.uci.edu/ml/machine-learning-databases/housing/housing.data')
There are a few changes in the skeleton code compared to the previous post:
- The class instantation defines a new dictionary modelDiscardD
- The modelDiscardD is set to an empty list for each model included in ModelSelectSet.
- The extraction of selected features is specified separately for each model inside the RegrModTrainTest and RegrModKFold
These changes allows you to select different features for different models when comparing the effects of feature selection across different models.
Feature Selection
A dataset with many independent variables is likely to include variables that have little or no relevance for explaining the variation in the target variable, or that are redundant. With many variables the risk for identifying random effects as true explanations also increases. Applying a feature selection prior to formulating a machinelearning model thus has several benefits:
- reduced the risk of identifying false explanation
- improves model robustness
- speeds up the modelling
The Scikit learn page on Feature selection introduces the feature selection alternative available in sklearn.
Variance threshold
A simple baseline method for feature selection is to look at the variance among the independent variables while disregarding the target. As the target is disregarded the method can be applied to all datasets. To test it import MinMaxScaler and VarianceThreshold from sklearn to your module.
from sklearn.preprocessing import MinMaxScaler
from sklearn.feature_selection import VarianceThreshold
As the features we have for predicting housing prices represent all kinds of units, they have widely varying ranges. You will thus use MinMaxScaler to rescale all the feature data to have the same range before applying the feature selection using VarianceThreshold .
Create the function VarianceSelector as part of the class RegressionModels. In the function all the feature data are first scaled to between 0 and 1, and then the threshold for accepting or discarding each feature is applied. The function then sets the discarded features as a list to all models defined to be used. The default threshold for discarding features is 0, which means that only features that have a constant value will be discarded. You can set any other threshold (between 0 and 1) when calling VarianceSelector.
def VarianceSelector(self,thresh=0):
#Initiate the MinMaxScale
scaler = MinMaxScaler()
#Print the scaling function
print ('Scaling function:'),(scaler.fit(self.X))
#Scale the data as defined by the scaler
Xscaled = scaler.transform(self.X)
#Initiate VarianceThreshold
select = VarianceThreshold(threshold=thresh)
#Fit the independent variables
select.fit(Xscaled)
#Get the selected features from get_support as a boolean list with True or False
selectedFeatures = select.get_support()
#Create a list to hold discarded columns
discardL = []
#print the selected features and their variance
print ('\nSelected features:')
for sf in range(len(selectedFeatures)):
if selectedFeatures[sf]:
print (' '),(self.columns[sf]), (select.variances_[sf])
else:
discardL.append([self.columnsX[sf],select.variances_[sf]])
print ('\nDiscarded features:')
for sf in discardL:
print (' '),(sf[0]), (sf[1])
#Redo the list of discarded features to only contain the column name
discardL = [item[0] for item in discardL]
#Set the list of discarded features to all defined models
for key in self.modD:
self.modelDiscardD[key] = discardL
To call the VarianceTest function, add the following line at the end of the __main__ section:
regmods.VarianceSelector(0.1)
If you run the module with thresh set to 0.1 (as in the example above), only two features (‘RAD’ and ‘TAX’) will be selected.
Selected features:
RAD 0.143036921113
TAX 0.103245089155
Discarded features:
CRIM 0.0093284136076
ZN 0.0542861839741
INDUS 0.0631168109761
CHAS 0.0643854770423
NOX 0.0567371966021
RM 0.0180885498782
AGE 0.0838733649487
DIS 0.0365929549544
PTRATIO 0.0529394103248
B 0.0528898018633
LSTAT 0.0387516350341
Test how an ordinary least square (OLS) regression performs with the selected features, by adding the following lines to the __main__ section:
#define the models to use
regmods.modD = {}
regmods.modD['OLS'] = {}
#Invoke the models
regmods.ModelSelectSet()
#Run the feature selection process
regmods.VarianceSelector(0.1)
#Run the modelling
regmods.RegrModTrainTest()
Run the module. All the models you defined in the __main__ section will go into the loop in RegrModTrainTest.
for m in self.models:
#Retrieve the model name and the model itself
name,mod = m
#Remove the features listed in the modelDiscarD
self.ExtractDf(self.modelDiscardD[name])
The function ExtractDf extracts only the selected features for the class variable X (self.X) used in the actual regression.
You can compare the results, shown in the figure below, with the results when using all the features (the first figure in the previous post).
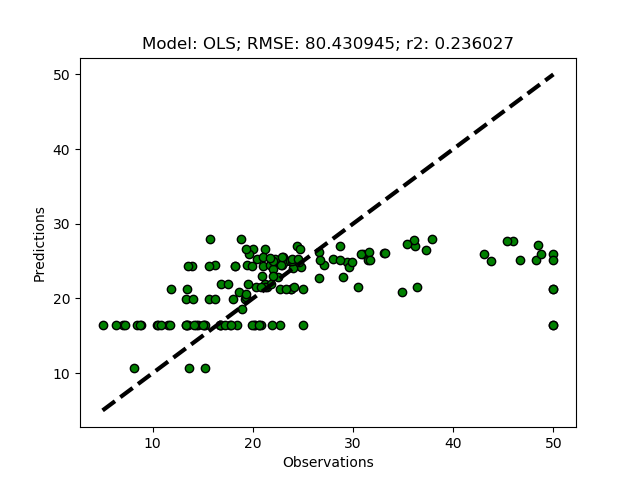
If you lower the threshold, more features will be selected, and if you set it to 0 (the default), all features will be selected.
Univariate feature selection
Univariate feature selection identifies the independent features that have the strongest relationship with the target feature. The univariate feature selectors available in sklearn are summarised on the Scikit learn page on Feature selection. In this post you are going to implement feature regression (f_regression) statistical test in the Python module.
Import the required functions from sklearn.
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_regression
Then create the function UnivariateSelector.
def UnivariateSelector(self,kselect):
#Initiate SelectKBest using f_regression
select = SelectKBest(score_func=f_regression, k=kselect)
#Fit the independent variables
select.fit(self.X, self.y)
#Get the selected features from get_support as a boolean list with True or False
selectedFeatures = select.get_support()
#Create a list to hold discarded columns
discardL = []
#print the selected features and their variance
print ('\nSelected features:')
for sf in range(len(selectedFeatures)):
if selectedFeatures[sf]:
print (' '),(self.columns[sf]), (select.pvalues_ [sf])
else:
discardL.append([self.columnsX[sf],select.pvalues_ [sf]])
print ('\nDiscarded features:')
for sf in discardL:
print (' '),(sf[0]), (sf[1])
#Redo the list of discarded features to only contain the column names
discardL = [item[0] for item in discardL]
#Set the list of discarded features to all defined models
for key in self.modD:
self.modelDiscardD[key] = discardL
The function UnivariateSelector requires that you give the number of features you want to select, there is no default number set. To try UnivariateSelector out, add the call to the function after the call to VarianceSelector and comment out the latter. The code snippet below shows the last parts of __main__ section, with UnivariateSelector set to select 2 features.
#regmods.VarianceSelector(0.1)
regmods.UnivariateSelector(2)
regmods.RegrModTrainTest()
The univariate selector picks out 2 other features (‘RM’ and ‘LSTAT’), and the prediction is much better compared to when selecting features using the variance and disregarding the target.
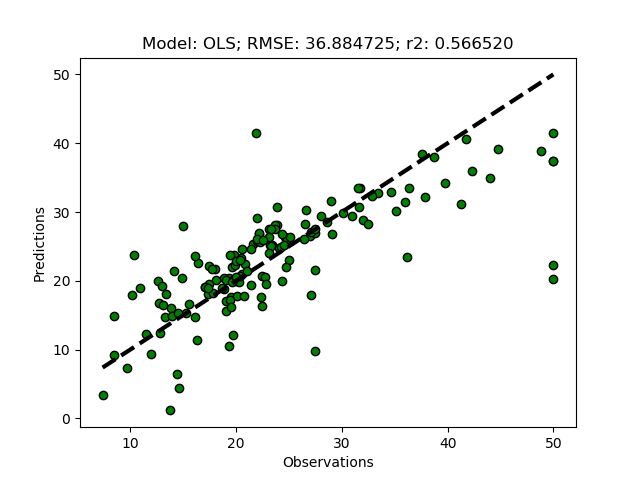
Recursive Feature Elimination (RFE)
RFE is a more powerful feature selector, relating feature importance to model performance. RFE works by recursively removing features and constructing a model on the surviving features. More details on the Scikit learn page on Feature selection.
Add the RFE function from sklearn.
from sklearn.feature_selection import RFE
Then create a general function for feature selection using FRE (RFESelector).
def RFESelector(self,kselect):
for mod in self.models:
select = RFE(estimator=mod[1], n_features_to_select=kselect, step=1)
select.fit(self.X, self.y)
selectedFeatures = select.get_support()
#Create a list to hold discarded columns
discardL = []
#print the selected features and their variance
print ('\nRegressor: %(m)s' %{'m':mod[0]})
print ('\nSelected features:')
for sf in range(len(selectedFeatures)):
if selectedFeatures[sf]:
print (' '),(self.columns[sf])
else:
discardL.append(self.columnsX[sf])
print ('\nDiscarded features:')
for sf in discardL:
print (' '),(sf)
self.modelDiscardD[mod[0]] = discardL
Not all models can be used for RFE feature selection. The recursive selector requires that a score is returned in each loop, and and not all models expose “coef_” or “feature_importances_” (as the error message will say if you try). That excludes for instance the KNeighborsRegressor. However, it is not required to use the same model for feature selection and prediction. And nothing prevents you from using one model for feature selection using RFE, and another model for predicting the target from the surviving features.
Add the models you want to test and then call RFESelector from the __main__ section. In the code below I also switched from using the train+test approach (RegrModTrainTest) to iterative cross validation (RegrModKFold) for model testing, as discuseed in the previous post. The number of features to find is set to 5 for all models (kselect = 5)
#define the models to use
regmods.modD = {}
regmods.modD['OLS'] = {}
regmods.modD['DecTreeRegr'] = {}
regmods.modD['RandForRegr'] = {'n_estimators':30}
regmods.modD['SVR'] = {'kernel':'linear','C':1.5,'epsilon':0.05}
#Invoke the models
regmods.ModelSelectSet()
#Run the feature selection process
#regmods.VarianceSelector(0.1)
#regmods.UnivariateSelector(2)
kselect = 5
regmods.RFESelector(kselect)
print ('Summary discarded features')
for key in regmods.modelDiscardD:
print ( ' %s: %s' %(key, regmods.modelDiscardD[key]) )
#regmods.RegrModTrainTest()
regmods.RegrModKFold()
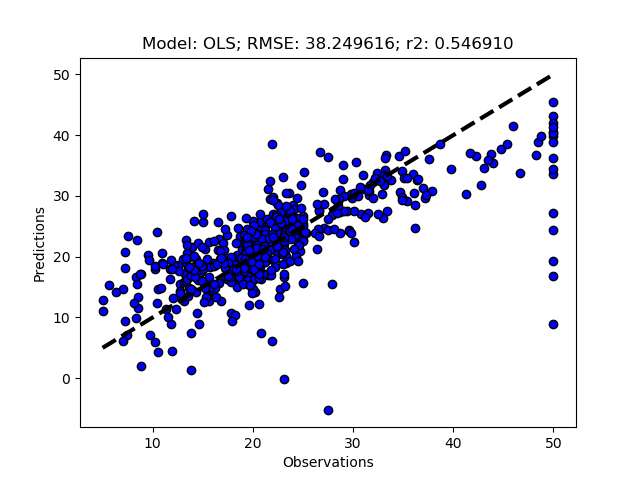
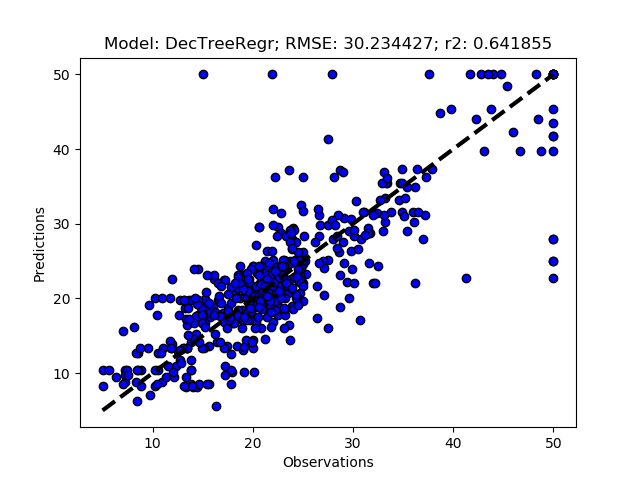
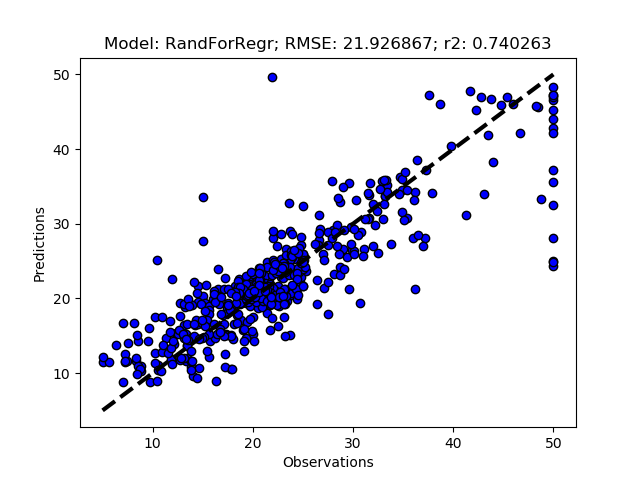
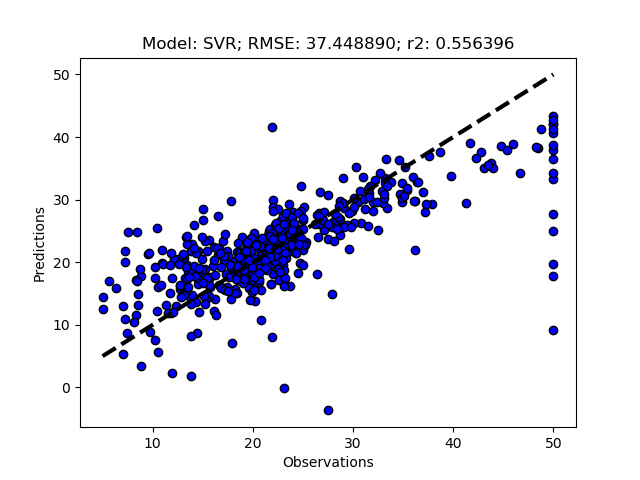
The complete Python module is availabe on Karttur’s repository on Github.
Resources
Scikit learn page on Feature Selection
Feature Selection in Python with Scikit-Learn by Jason Brownlee
Completed python module on GitHub.