Contents - Introduction - Prerequisits - Python module skeleton - Model hyper-parameters - KNeighborsRegressor - DecisionTreeRegressor - Support Vector Machine Regressor (SVR) - RandomForestRegressor - Tuning the parameters - Report function - Model parameterization - KNeighborsRegressor - DecisionTreeRegressor - Support Vector Machine Regressor (SVR) - RandomForestRegressor - Model Predictions - Resources
Introduction
If you have followed Karttur’s blog posts on machine learning, you have learnt to apply different regressors for predicting continuous variables, and how to discriminate among the independent variables. But so far you have either only used the default parameters (called hyper-parameters in Scikit learn) defining the regressors, or tested a few other settings. So while the regressors that you applied have used training data (or folded cross validation) for the internal parameters linking the independent variables to the target feature, the hyper-parameters defining how this is done have been fixed.
The Scikit learn package includes two modules for tuning (or optimizing) the hyper-parameter settings, Exhaustive Grid Search (GridSearchCV) and Randomized search (RandomizedSearchCV). You get an introduction to both at the Scikit learn page on tuning the hyper-parameters.
As in the previous Karttur posts on machine learning, this post will evolve around creating a Python module (.py file) for applying the tuning of hyper-parameters for different regressors.
You can copy and paste the code in the post, but you might have to edit some parts: indentations does not always line up, quotations marks can be of different kinds and are not always correct when published/printed as html text, and the same also happens with underscores and slashes.
Prerequisits
The prerequisites are the same as in the previous posts in this series: a Python environment with numpy, pandas, sklearn (Scikit learn) and matplotlib installed.
Python module skeleton
As in the previous posts on machine learning, we start by a Python module (.py file) skeleton that will be used as a vehicle for developing a complete sklearn hyper-parameter selection module. The skeleton code is hidden under the button.
Model hyper-parameters
All sklearn models have a different suite of hyper-parameters that can be set. These parameters can be of four types:
- Integers
- Real (or float)
- Lists of alternatives
- Boolean (True or False)
When tuning hyper-parameters the first thing to decide is which parameters to tune. You can find out which hyper-parameters that can be passed to all sklearn models in the Scikit pages for each regressor. But you can also get them as a dictionary in Python, and you will explore them further down. Create the function RandomTuningParams, under the class RegressionModels. At first you will only use the function for exploring the parameters to set, the actual parameter settings for tuning will be added later.
def RandomTuningParams(self):
# specify parameters and distributions to sample from
for m in self.models:
name,mod = m
print ('name'), (name), (mod.get_params())
KNeighborsRegressor
To explore the hyper-parameters of Scikit learn regressors, define the models you want to explore, invoke them, and call the Tuningparameters function to see the hyper-parameters. The first example is for exploring the parameters for KNeighborsRegressor that is abbreviated ‘KnnRegr’ when added to the modD dictionary. Add the lines below to the __main__ section.
regmods.modD = {}
regmods.modD['KnnRegr'] = {}
#Invoke the models
regmods.ModelSelectSet()
#Tuning parameters
regmods.RandomTuningParams(11)
Run the module, and check the listed hyper-parameters and their default values.
name KnnRegr {'n_neighbors': 5, 'n_jobs': 1, 'algorithm': 'auto', 'metric': 'minkowski',
'metric_params': None, 'p': 2, 'weights': 'uniform', 'leaf_size': 30}
DecisionTreeRegressor
Add the ‘DecTreeRegr’ (DecisionTreeRegressor) regressor to the modD dictionary
regmods.modD['DecTreeRegr'] = {}
The model hyper-parameters for the DecisionTreeRegressor:
name DecTreeRegr {'presort': False, 'splitter': 'best', 'min_impurity_decrease': 0.0, 'max_leaf_nodes': None,
'min_samples_leaf': 1, 'min_samples_split': 2, 'min_weight_fraction_leaf': 0.0, 'criterion': 'mse',
'random_state': None, 'min_impurity_split': None, 'max_features': None, 'max_depth': None}
Support Vector Machine Regressor (SVR)
Add the ‘SVR’ (SVR) to the modD dictionary
regmods.modD['SVR'] = {}
The model hyper-parameters for the SVR:
name SVR {'kernel': 'rbf', 'C': 1.0, 'verbose': False, 'degree': 3, 'epsilon': 0.1, 'shrinking': True,
'max_iter': -1, 'tol': 0.001, 'cache_size': 200, 'coef0': 0.0, 'gamma': 'auto'}
RandomForestRegressor
Add the ‘RandForRegr’ (RandomForestRegressor) regressor to the modD dictionary
regmods.modD['RandForRegr'] = {}
The model hyper-parameters for the RandomForestRegressor:
name RandForRegr {'warm_start': False, 'oob_score': False, 'n_jobs': 1, 'min_impurity_decrease': 0.0,
'verbose': 0, 'max_leaf_nodes': None, 'bootstrap': True, 'min_samples_leaf': 1, 'n_estimators': 10,
'min_samples_split': 2, 'min_weight_fraction_leaf': 0.0, 'criterion': 'mse', 'random_state': None,
'min_impurity_split': None, 'max_features': 'auto', 'max_depth': None}
Tuning the parameters
Before you can use the module for tuning the hyper-parameters, you must create a reporting function, and the functions for setting the hyper-parameters to tune.
Report function
Add the reporting function (ReportSearch).
def ReportSearch(self, results, n_top=3):
for i in range(1, n_top + 1):
candidates = np.flatnonzero(results['rank_test_score'] == i)
for candidate in candidates:
print("Model with rank: {0}".format(i))
print("Mean validation score: {0:.3f} (std: {1:.3f})".format(
results['mean_test_score'][candidate],
results['std_test_score'][candidate]))
print("Parameters: {0}".format(results['params'][candidate]))
print("")
Randomized tuning
As I wanted to try both randomized and exhaustive tuning, I opted for creating a separate function for each method. The function RandomTuning invokes the randomized tuning search, prints the results of the tuning search, and then also sets the highest ranked hyper-parameter setting as the parameters for each model (by updating the modD dictionary).
def RandomTuning(self, fraction=0.5, nIterSearch=6, n_top=3):
#Randomized search
for m in self.models:
#Retrieve the model name and the model itself
name,mod = m
print name, self.paramDist[name]
search = RandomizedSearchCV(mod, param_distributions=self.paramDist[name],
n_iter=nIterSearch)
X_train, X_test, y_train, y_test = model_selection.train_test_split(self.X, self.y, test_size=(1-fraction))
search.fit(X_train, y_train)
self.ReportSearch(search.cv_results_,n_top)
#Retrieve the top ranked tuning
best = np.flatnonzero(search.cv_results_['rank_test_score'] == 1)
tunedModD=search.cv_results_['params'][best[0]]
#Append any initial modD hyper-parameter definition
for key in self.modD[name]:
tunedModD[key] = self.modD[name][key]
regmods.modD[name] = tunedModD
Without setting any parameters, the tuning search for each model is defaulted to use half of the dataset (parameter fraction=0.5) for the tuning, 6 iterations (parameter _nIterSearch=6), and to print out the top 3 results (parameter n_top=3). For each regressor, the hyper-parameters for the best tuning are retrieved. If the regressor model had any initial hyper-parameters set in the modD dictionary they are added, and the tuned hyper-parameters are then set as parameter+value pairs in modD.
The RandomTuning function uses the Scikit learn randomized tuning function RandomizedSearchCV, that you must add to the imports at the beginning of the module. Then you will also need to import functions for creating ranges of randomized numbers.
from scipy.stats import randint as sp_randint
from scipy.stats import uniform as sp_randreal
from sklearn.model_selection import RandomizedSearchCV
You also have to call RandomTuning from the __main__ section
regmods.RandomTuning()
If you want to increase the search iterations to 12, and the print out the top 6 results, but keep the fraction of the dataset at 0.5, add that to the call.
regmods.RandomTuning(0.5,12,6)
Then you have to create the values for the parameter param_distributions used in RandomTuning (param_distributions=self.paramDist). The values to send to param_distributions are defined in the variable self.paramDist, and defines both which hyper-parameters to tune, and what values each hyper-parameter is allowed to take. You have to look at the individual Scikit learn pages to get a grip on the hyper-parameters you want to tune, and what ranges/alternatives you can/want to set. The principle for setting the ranges/parameters differs for the different types of parameters.
- Integers: sp_randint(min, max) or predefined set (i, j, k, …)
- Real: sp_randreal(min, max) or predefined set (i.j, k.l, m.n, …)
- Alternatives: [‘alt1’, ‘alt2’, ‘alt3’, …]
- Boolean: [True, False]
KNeighborsRegressor randomized tuning
The KNeighborsRegressor (‘KnnRegr’) regressor has fewer hyper-parameters compared to the other non-linear regressors used in here (see above). The code snippet below defines the randomized tuning for the ‘KnnRegr’ hyper-parameters n_neighbors, leaf_size, weight, p and algorithm.
def RandomTuningParams(self):
# specify parameters and distributions to sample from
for m in self.models:
name,mod = m
print ('name'), (name), (mod.get_params().keys())
if name == 'KnnRegr':
self.paramDist = {"n_neighbors": sp_randint(4, 12),
'leaf_size': sp_randint(10, 50),
'weights': ('uniform','distance'),
'p': (1,2),
'algorithm': ('auto','ball_tree', 'kd_tree', 'brute')}
Run the Pyton module to get the tuned hyper-parameter for ‘KnnRegr’. As the process uses a randomizer, the results varies each time you run it, but should resemble the results shown below.
Model with rank: 1
Mean validation score: 0.373 (std: 0.158)
Parameters: {'n_neighbors': 7, 'weights': 'distance', 'leaf_size': 28, 'algorithm': 'auto', 'p': 1}
Model with rank: 2
Mean validation score: 0.371 (std: 0.136)
Parameters: {'n_neighbors': 10, 'weights': 'distance', 'leaf_size': 24, 'algorithm': 'ball_tree', 'p': 1}
Model with rank: 2
Mean validation score: 0.371 (std: 0.136)
Parameters: {'n_neighbors': 10, 'weights': 'distance', 'leaf_size': 15, 'algorithm': 'auto', 'p': 1}
Transferring the best result from the tuning above (“Model with rank: 1”) to the model, the full model hyper-parameter settings for the tuned ‘KnnRegr’ is shwon below.
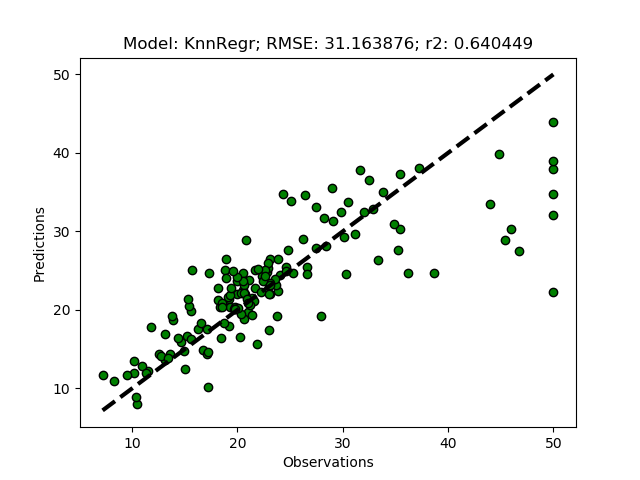
name KnnRegr {'n_neighbors': 7, 'n_jobs': 1, 'algorithm': 'auto', 'metric': 'minkowski', 'metric_params': None, 'p': 1, 'weights': 'distance', 'leaf_size': 28}
DecisionTreeRegressor randomized tuning
For the DecisionTreeRegressor (‘KnnRegr’) regressor I opted for tuning max_depth, min_samples_split and min_samples_leaf.
elif name =='DecTreeRegr':
self.paramDist[name] = {"max_depth": [3, None],
"min_samples_split": sp_randint(2, 6),
"min_samples_leaf": sp_randint(1, 4)}
With the following results:
Model with rank: 1
Mean validation score: 0.479 (std: 0.162)
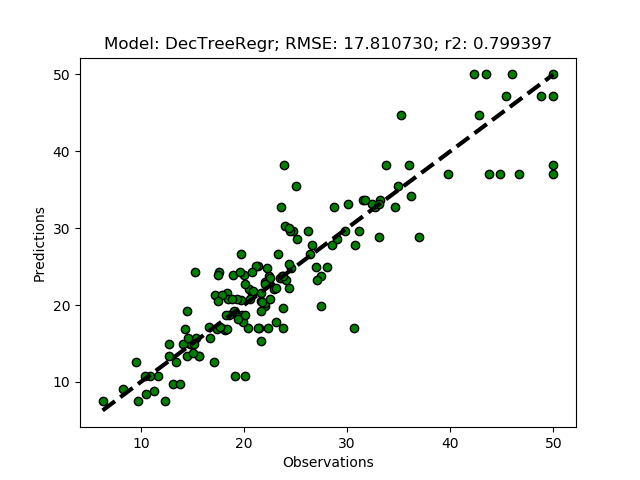
Parameters: {'min_samples_split': 4, 'max_depth': None, 'min_samples_leaf': 3}
Model with rank: 2
Mean validation score: 0.367 (std: 0.206)
Parameters: {'min_samples_split': 3, 'max_depth': 3, 'min_samples_leaf': 3}
Model with rank: 3
Mean validation score: 0.367 (std: 0.206)
Parameters: {'min_samples_split': 5, 'max_depth': 3, 'min_samples_leaf': 3}
SVR randomized tuning
For the SVR (‘SVR’) regressor I opted for tuning kernel, epsilon, and C. Rather than using a randomizer I hardcoded the values open for epsilon and C (with more values the processing takes a very long time).
elif name =='SVR':
self.paramDist[name] = {"kernel": ['linear', 'rbf'],
"epsilon": (0.05, 0.1, 0.2),
"C": (1, 2, 5, 10)}
With the following results:
Model with rank: 1
Mean validation score: 0.724 (std: 0.083)
Parameters: {'kernel': 'linear', 'C': 1, 'epsilon': 0.2}
Model with rank: 2
Mean validation score: 0.714 (std: 0.126)
Parameters: {'kernel': 'linear', 'C': 5, 'epsilon': 0.1}
Model with rank: 3
Mean validation score: 0.041 (std: 0.021)
Parameters: {'kernel': 'rbf', 'C': 10, 'epsilon': 0.05}
Note the large difference in validation score between highest ranked ranked models (‘linear’ kernel), and the 3rd model with the ‘rbf’ kernel. The latter also has the largest allowed value of the C hyper-parameter.
RandomForestRegressor randomized tuning
For the RandomForestRegressor (‘RandForRegr’) regressor I opted for tuning max_depth, n_estimators, max_features, min_samples_split , min_samples_leaf and bootstrap.
elif name =='RandForRegr':
self.paramDist = {"max_depth": [3, None],
"n_estimators": sp_randint(10, 50),
"max_features": sp_randint(1, max_features),
"min_samples_split": sp_randint(2, up_samples_split),
"min_samples_leaf": sp_randint(1, up_samples_leaf),
"bootstrap": [True,False]}
With the following results:
Model with rank: 1
Mean validation score: 0.744 (std: 0.075)
Parameters: {'bootstrap': False, 'min_samples_leaf': 3, 'n_estimators': 17, 'min_samples_split': 2, 'max_features': 9, 'max_depth': None}
Model with rank: 2
Mean validation score: 0.727 (std: 0.116)
Parameters: {'bootstrap': True, 'min_samples_leaf': 2, 'n_estimators': 31, 'min_samples_split': 4, 'max_features': 3, 'max_depth': None}
Model with rank: 3
Mean validation score: 0.727 (std: 0.073)
Parameters: {'bootstrap': False, 'min_samples_leaf': 4, 'n_estimators': 13, 'min_samples_split': 5, 'max_features': 6, 'max_depth': 3}
Randomized Model Predictions
The model setup, using the modD dictionary, allows the tuned hyper-parameters to be set directly to each model. The hyper-parameters of each regressor are updated as part of the function RandomTuning. To invoke the tuned hyper-parameters, you have to reset the models in the __main__ section, and then call either RegrModTrainTest or RegrModKFold function, or both, to run the tuned models for your dataset.
#Reset the models with the tuned hyper-parameters
regmods.ModelSelectSet()
#Run the models
regmods.RegrModTrainTest()
regmods.RegrModKFold()
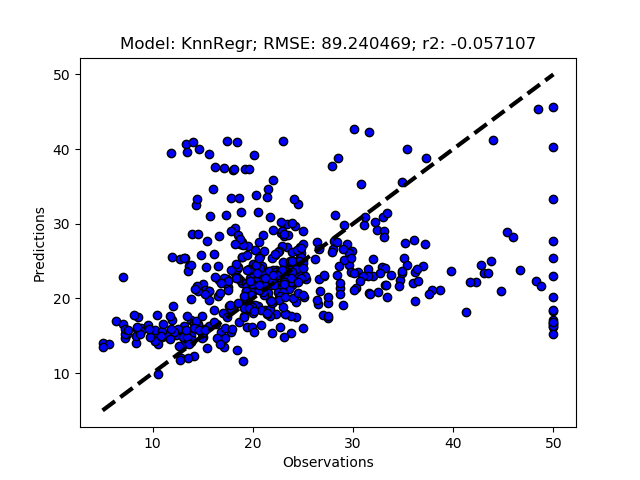
Randomized tuning results
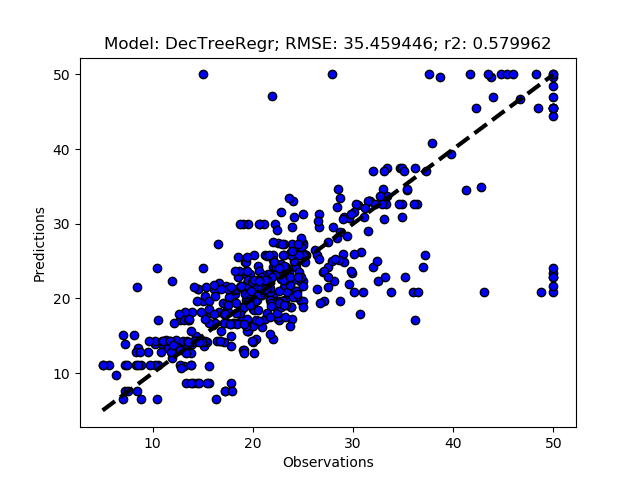
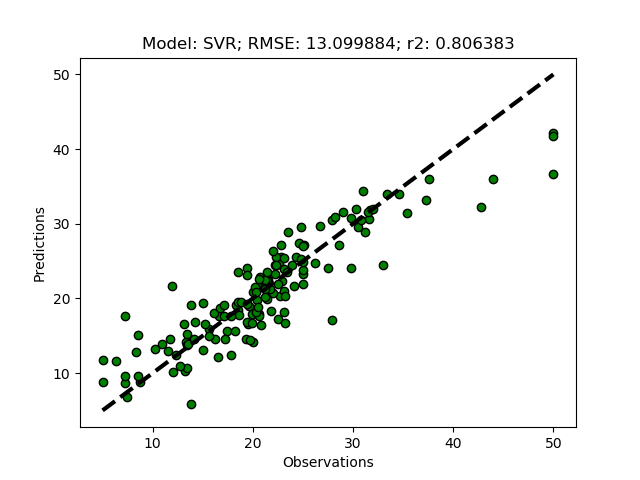
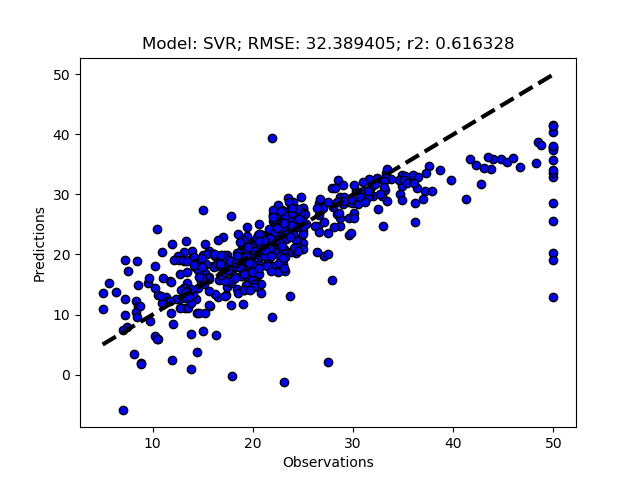
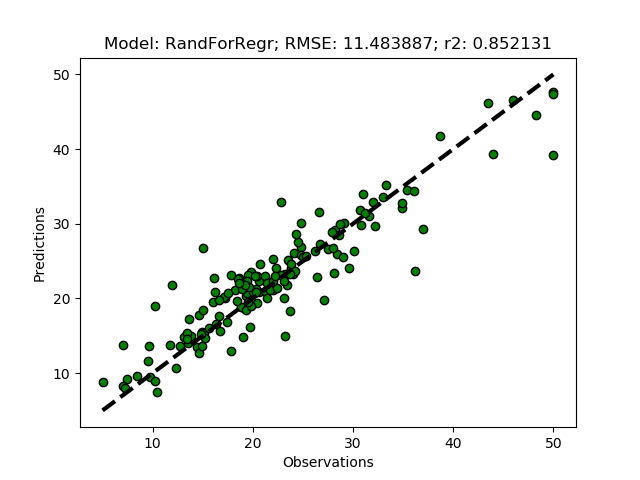
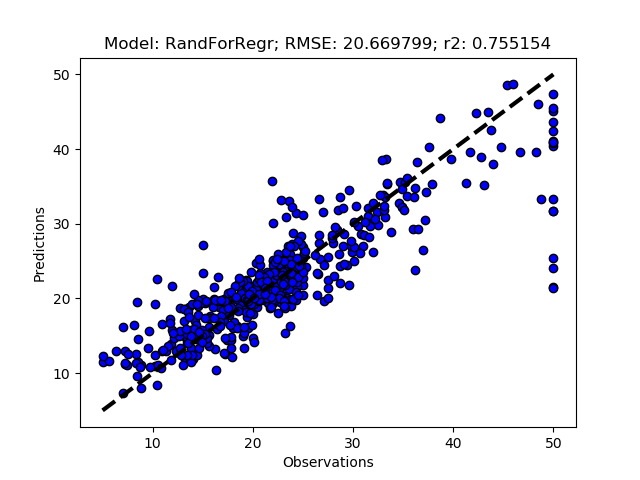
Exhaustive tuning
The exhaustive tuning (or exhaustive grid search) is provided by the Scikit learn function GridSearchCV. The function exhaustively generates candidates from a grid of hyper-parameter values specified with the param_grid parameter. Compared to the randomized grid search, you can specify the search space in more detail, but you need to narrow the search space down as the processes otherwise will take long. If your randomized tuning indicates that a hyper-parameter value can be set to a constant value that is not the default value, it is better to define that particular hyper-parameter in the initial model definition (modD) and omit it from the tuning search. If it is the default value of the hyper-parameter that can be held constant, all you have to do is to omit it from tuning search.
Import the GridSearchCV at the beginning of the module.
from sklearn.model_selection import GridSearchCV
And create the function ExhaustiveTuning under the class RegressionModels.
def ExhaustiveTuning(self, fraction=0.5, n_top=3):
# run exhaustive search
for m in self.models:
#Retrieve the model name and the model itself
name,mod = m
print name, self.paramGrid[name]
search = GridSearchCV(mod, param_grid=self.paramGrid[name])
X_train, X_test, y_train, y_test = model_selection.train_test_split(self.X, self.y, test_size=(1-fraction))
search.fit(X_train, y_train)
self.ReportSearch(search.cv_results_,n_top)
#Append items from the initial modD dictionary
best = np.flatnonzero(search.cv_results_['rank_test_score'] == 1)
tuneModD=search.cv_results_['params'][best[0]]
#Set the highest ranked hyper-parameter
for key in self.modD[name]:
tuneModD[key] = self.modD[name][key]
regmods.modD[name] = tuneModD
When setting the exhaustive tunings below, I have glanced as the top ranked results from the randomized tuning for each regressor. For some regressor models I also chose to set some of the tuned hyper-parameters from the randomized tuning search as initial hyper-parameters and omit them from the exhaustive tuning search.
KNeighborsRegressor exhaustive tuning
Add the function for setting the exhaustive search tuning parameters for each model to test. The code also contains the search setting for ‘KnnRegr’.
def ExhaustiveTuningParams(self):
# specify parameters and distributions to sample from
self.paramGrid = {}
for m in self.models:
name,mod = m
print ('name'), (name), (mod.get_params())
if name == 'KnnRegr':
self.paramGrid[name] = [{"n_neighbors": [6,7,8,9,10],
'algorithm': ('ball_tree', 'kd_tree'),
'leaf_size': [15,20,25,30,35]},
{"n_neighbors": [6,7,8,9,10],
'algorithm': ('auto','brute')}
]
The hyper-parameter leaf_size in ‘KnnRegr’ only has relevance when the hyper-parameter algorithm is set either to ball_tree or kd_tree. The search is thus divided into two blocks (each defined as a dictionary), the first block for ball_tree and kd_tree also includes leaf_size, whereas the second block (for the algorithms auto and brute) does not. In the randomized tuning search for ‘KnnRegr’, the three top ranked results all had the hyper-parameter p (power parameter for the [default] Minkowski metric) value of 1, whereas the default value is 2. Also the hyper-parameter weights have a constant value (distance) in the top ranked randomized tunings, and this is also not the default value. When initially formulating the ‘KnnRegr’ model (in the __main__ section), I thus set the hyper-parameters p to 1, and weights to ‘distance’ and omit them from the exhaustive tuning.
regmods.modD['KnnRegr'] = {'weights':'distance','p':1}
The results from the exhaustive search with these settings are similar to the results from the randomized search. And the regressors appears to be insensitive to most of the hyper-parameters, as indicated from the five equally ranked parameter settings below.
Model with rank: 1
Mean validation score: 0.620 (std: 0.024)
Parameters: {'n_neighbors': 6, 'leaf_size': 15, 'algorithm': 'ball_tree'}
Model with rank: 1
Mean validation score: 0.620 (std: 0.024)
Parameters: {'n_neighbors': 6, 'leaf_size': 25, 'algorithm': 'ball_tree'}
Model with rank: 1
Mean validation score: 0.620 (std: 0.024)
Parameters: {'n_neighbors': 6, 'leaf_size': 15, 'algorithm': 'kd_tree'}
Model with rank: 1
Mean validation score: 0.620 (std: 0.024)
Parameters: {'n_neighbors': 6, 'algorithm': 'auto'}
Model with rank: 1
Mean validation score: 0.620 (std: 0.024)
Parameters: {'n_neighbors': 6, 'algorithm': 'brute'}
DecisionTreeRegressor exhaustive tuning
The ‘DecTreeRegr’ regressor does not have any hyper-parameter that is parameterized using a second hyper-parameter, and there is thus only a single search grid block for the exhaustive search.
elif name =='DecTreeRegr':
self.paramGrid =[{
"min_samples_split": [2,3,4,5,6],
"min_samples_leaf": [1,2,3,4]}]
Model with rank: 1
Mean validation score: 0.783 (std: 0.008)
Parameters: {'min_samples_split': 6, 'min_samples_leaf': 1}
Model with rank: 2
Mean validation score: 0.776 (std: 0.047)
Parameters: {'min_samples_split': 3, 'min_samples_leaf': 1}
Model with rank: 3
Mean validation score: 0.754 (std: 0.038)
Parameters: {'min_samples_split': 5, 'min_samples_leaf': 1}
SVR exhaustive tuning
The SVR regressor can be set with different kernels (hyper-parameter kernel), with different additional hyper-parameters used for defining the behaviour of different kernels. The SVR regressor is computationally demanding, and you must be careful when setting the tuning options unless you have a very powerful machine, or lots of time (or a small dataset).
elif name =='SVR':
self.paramGrid = [{"kernel": ['linear'],
"epsilon": (0.05, 0.1, 0.2),
"C": (1, 10, 100)},
{"kernel": ['rbf'],
'gamma': [0.001, 0.0001],
"epsilon": (0.05, 0.1, 0.2),
"C": (1, 10, 100)},
{"kernel": ['poly'],
'gamma': [0.001, 0.0001],
'degree':[2,3],
"epsilon": (0.05, 0.1, 0.2),
"C": (0.5, 1, 5, 10, 100)}]
All the highest ranked models have the hyper paramteer kernel set to ‘linear’, with results insensitive to both the hyper-parameters C and epsilon within the ranges set in the exhaustive tuning search.
Model with rank: 1
Mean validation score: 0.604 (std: 0.020)
Parameters: {'epsilon': 0.2, 'C': 1, 'kernel': 'linear'}
Model with rank: 2
Mean validation score: 0.602 (std: 0.021)
Parameters: {'epsilon': 0.2, 'C': 2, 'kernel': 'linear'}
Model with rank: 3
Mean validation score: 0.600 (std: 0.016)
Parameters: {'epsilon': 0.1, 'C': 1, 'kernel': 'linear'}
RandomForestRegressor exhaustive tuning
The ‘RandForRegr’ has plenty of hyper-parameters that can be set. I opted for only tuning a few, including n_estimators, min_samples_split, min_samples_leaf and bootstrap.
elif name =='RandForRegr':
self.paramGrid[name] = [{
"n_estimators": (20,30),
"min_samples_split": (2, 3, 4, 5),
"min_samples_leaf": (2, 3, 4),
"bootstrap": [True,False]}]
The results of the random forest regressor varies between different runs. This happens because the initial branching (how data is split in the growing trees) determines later branching and the forest can look vary different in different runs.
Model with rank: 1
Mean validation score: 0.848 (std: 0.071)
Parameters: {'min_samples_split': 2, 'n_estimators': 30, 'bootstrap': True, 'min_samples_leaf': 2}
Model with rank: 2
Mean validation score: 0.848 (std: 0.071)
Parameters: {'min_samples_split': 3, 'n_estimators': 30, 'bootstrap': True, 'min_samples_leaf': 2}
Model with rank: 3
Mean validation score: 0.842 (std: 0.063)
Parameters: {'min_samples_split': 4, 'n_estimators': 20, 'bootstrap': True, 'min_samples_leaf': 2}
Exhausted tuning results
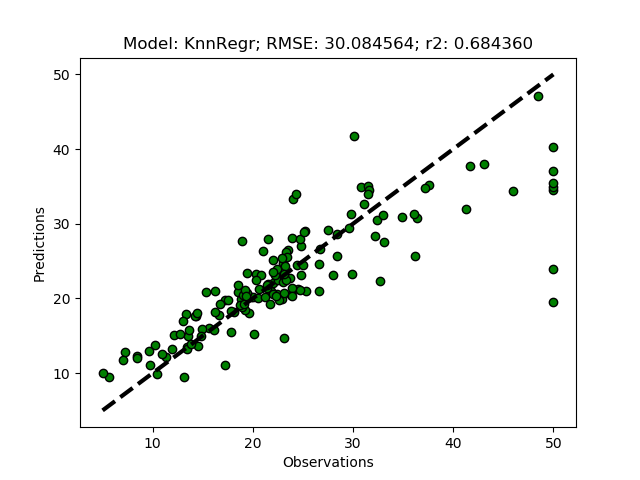
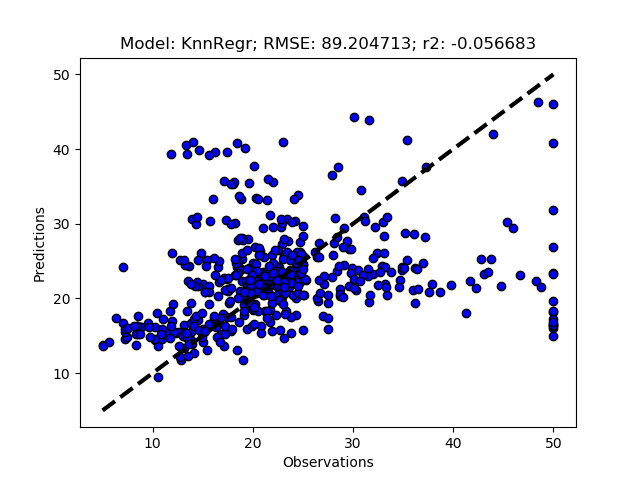
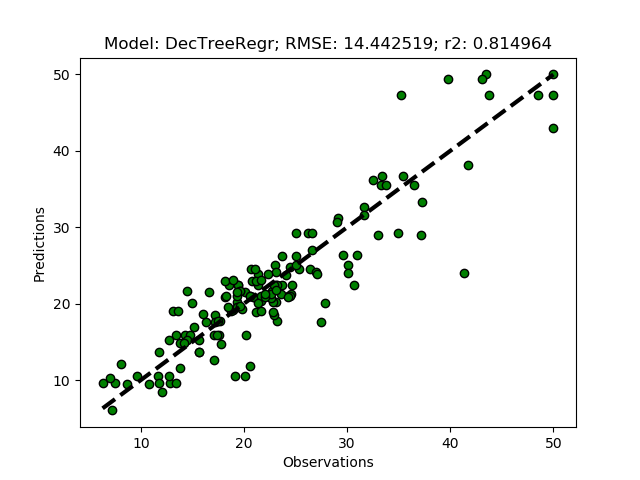
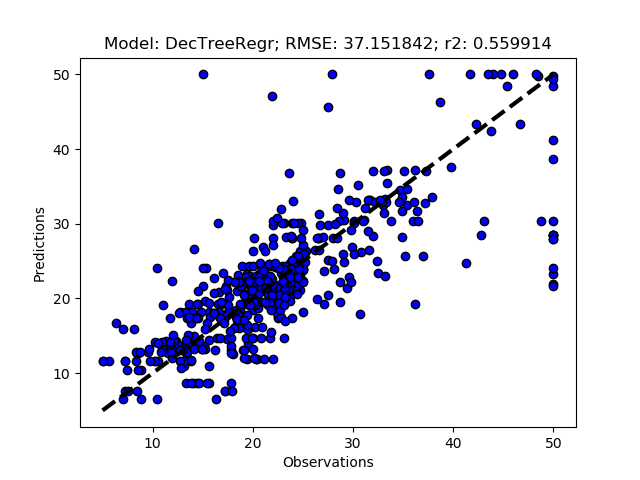
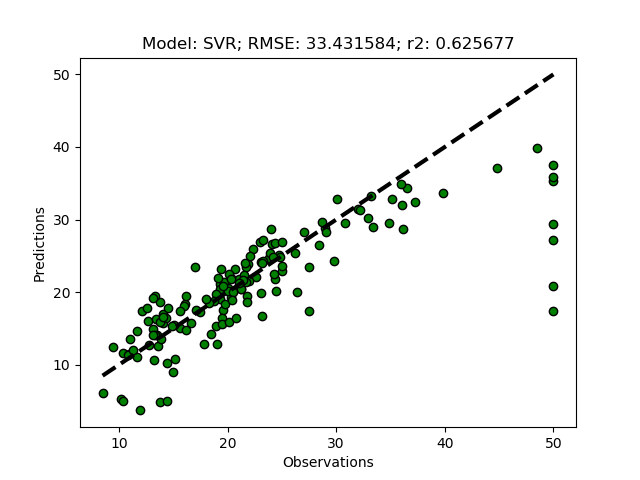
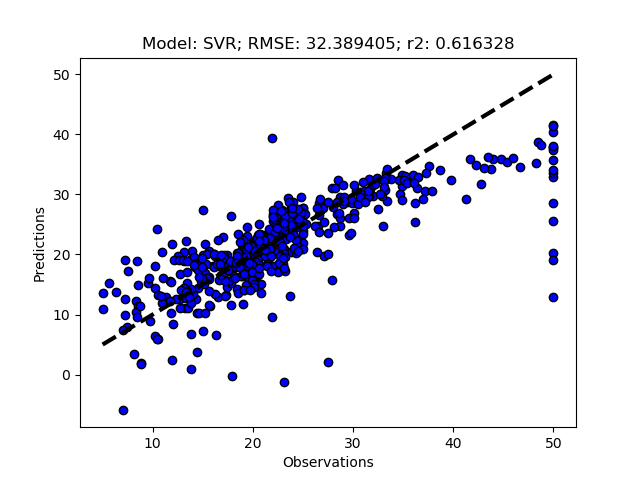
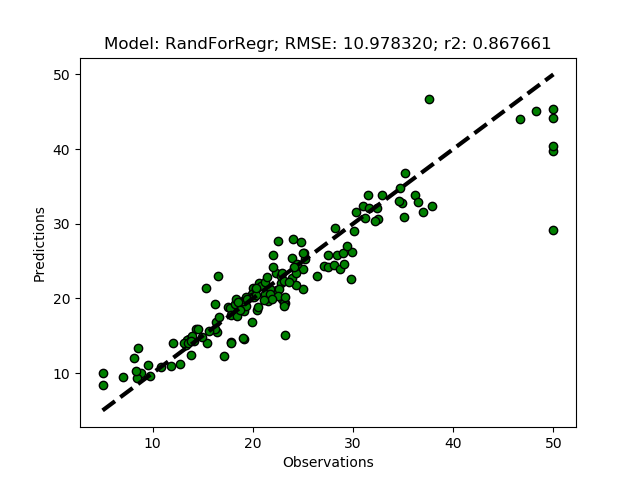
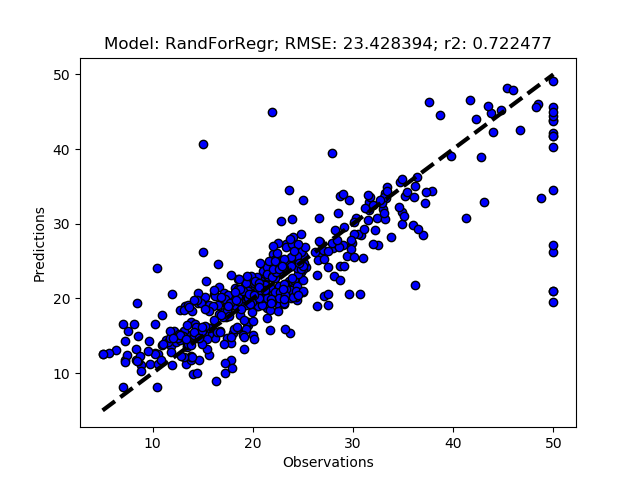
The complete Python module is availabe on Karttur’s repository on Github.
Resources
Tuning the hyper-parameters of an estimator, Scikit learn.
RandomizedSearchCV, Scikit learn.
GridSearchCV, Scikit learn.
What is the Difference Between a Parameter and a Hyperparameter? by Jason Brownlee
Completed python module on GitHub.