Contents - Introduction - Prerequisites - Module Skeleton - Pipeline - LarsCV feature selection and regression - Pipeline feature selection and regression - Emebbeding the pipeline in a cross validation search - A more general function - Resources
Introduction
If you followed Karttur’s series of tuturials on machine learning, you encountered a pipeline in the post on feature selection. This post dives into the construction of pipelines in Scikit learn.
You can copy and paste the code in the post, but you might have to edit some parts: indentations does not always line up, quotations marks can be of different kinds and are not always correct when published/printed as html text, and the same also happens with underscores and slashes.
The complete code is also available here.
Prerequisites
To follow the post you need a Python environment with numpy, pandas, sklearn (Scikit learn) and matplotlib installed.
Module Skeleton
The module skeleton code is under the button.
Pipeline
In a Scikit learn pipeline you can link different processes together. The requirement is that each process (except the last) must be able to transform the input and then send the transformed results to the next step. The transformation itself is automated, but not all functions expose the transform capacity. If you include a process that can not use transform , the pipeline causes a runtime error, and reports that the step does not expose transform capacity.
There are several advantages using pipelines, including eliminating memory leakage, more efficient processing, and less coding. Once you understand the concept.
In the next section you will set up a regression without a pipeline. To remove all random elements in the regression process, the calibration (fitting) and validation (prediction) will use the same data. In the following section you will set up exactly the same process, but using a pipeline. As the random effects are removed, the result should be exactly the same.
LarsCV feature selection and regression
In this post I chose to use the Least Angle Regression (Lars) regressor. Lars is a general linear regressor that resembles forward stepwise regression. It is particularly useful for high-dimensional data. LarsCV is an extension of Lars that includes a built-in cross validation (CV) function.
The function below (LarsCVFeatureSelect) contains three main parts:
- Feature selection using SelectFromModel
- Extraction of the selected features
- Application of the LarsCV regressor for fitting and predicting the target from the extracted feature
I have put in comments for all the steps in the function itself.
def LarsCVFeatureSelect(self, plot=False):
'''LarsCV feature selection and regression using the full dataset
'''
#Set the model to LarsCV
mod = LarsCV()
name = 'LarsCV'
#Define SelectFromModel using the model (LarsCV) and a threshold
select = SelectFromModel(mod, threshold="mean")
#Fit_transform the selection, it will return an X array only retaining features meeting the set threshold
X_new = select.fit_transform(self.X,self.y)
#Print the shape of the returned X_new array
print 'new', X_new.shape
#Get the number of features that was selected
nFeat = X_new.shape[1]
#Print the support of the selection (a list of boolean results for selected and discarded features)
print 'support',select.get_support()
#Create a dictionary from the original X columns and the boolean support list identifying the selected features
selectD = dict(zip(self.columnsX,select.get_support()))
#Extract the names of the selected features to a list
featureL = [item for item in selectD if selectD[item]]
#Print the selected features
print 'Selected features', featureL
#Fit the regression
fit = mod.fit(X_new, self.y)
#Print the parameters of the fitted regression
print 'fitted params', fit.get_params()
#Print the coefficiens of the fitted regression
print 'coefficients', fit.coef_
#Predict the target
predict = mod.predict(X_new)
#Prepare a message for printing out the results
title = ('Target: %(tar)s; Model: %(mod)s; n:%(n)s; RMSE: %(rmse)1.2f; r2: %(r2)1.2f' \
% {'tar':self.target,'mod':name,'n': nFeat, 'rmse':mean_squared_error(self.y, predict),'r2': r2_score(self.y, predict)} )
msg = ' Regression results\n %(t)s' %{'t':title}
#Print the result message
print (msg)
if plot:
self._PlotRegr(self.y, predict, title, color='maroon')
To run the function, call it from the __main__ section.
regmods.LarsCVFeatureSelect()
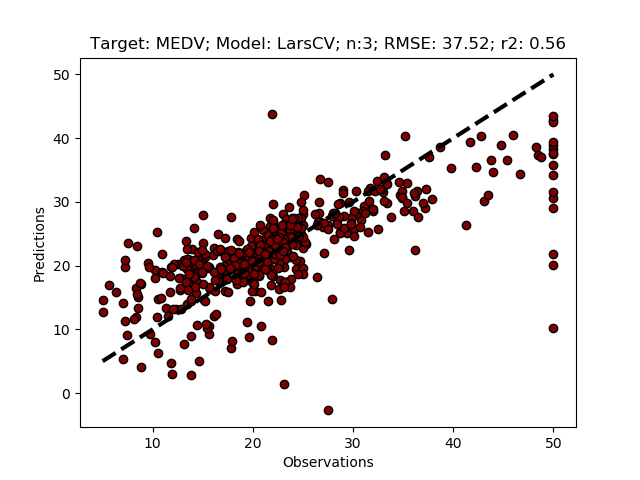
As noted above, the regression uses the same data for fitting and predicting the target. You will get exactly the same results every time you run the model. In the next section you will use a pipeline for linking the feature selection and the regression, and if it works should also give exactly the same results. If you change the call to the function to request a plot, the plotted image should be identical to the one below.
regmods.LarsCVFeatureSelect(True)
Pipeline feature selection and regression
In the function below, the feature selection and the regression are linked together in a pipeline. The function illustrates and explains how this is done, and how to retrieve the parameters and results from each step.
def LarsCVFeatureSelectPipeline(self, plot=False):
'''LarsCV feature selection and regression using the full dataset
'''
#Set the model to LarsCV
mod = LarsCV()
name = 'LarsCV'
#Define SelectFromModel using the model (LarsCV) and a threshold
select = SelectFromModel(mod, threshold="mean")
#Setup a pipeline linking the feature selection and the model
#At this stage the names of each step can be set to anything ('select' and 'regr')
pipeline = Pipeline([
('select',select),
('regr',mod)
])
#Fit the pipeline, each step will do a fit_transform and send the result to the next step
pipeline.fit(self.X, self.y)
#All the parameters from each step in the pipeline can be retrieved
#Print the support (list of selected and discarded) features from the 'select' step (step = 0)
print 'step 0 support',pipeline.steps[0][1].get_support()
#Each step can also be accessed using its name
print 'step select support', pipeline.named_steps['select'].get_support()
#Create a dictionary from the original X columns and the boolean support list identifying the selected features
selectD = dict(zip(self.columnsX,pipeline.named_steps['select'].get_support()))
#Extract the names of the selected features to a list
featureL = [item for item in selectD if selectD[item]]
#Print the selected features
print 'Selected features', featureL
#Get the number of features that was selected
nFeat = len(featureL)
#Print the parameters of the fitted regression
print 'fitted params', pipeline.steps[1][1].get_params()
#Print the coefficiens of the fitted regression
print 'coefficients', pipeline.named_steps['regr'].coef_
#Predict the target
predict = pipeline.predict(self.X)
#Prepare a message for printing out the results
title = ('Target: %(tar)s; Model: %(mod)s; n:%(n)s; RMSE: %(rmse)1.2f; r2: %(r2)1.2f' \
% {'tar':self.target,'mod':name,'n': nFeat, 'rmse':mean_squared_error(self.y, predict),'r2': r2_score(self.y, predict)} )
msg = ' Regression results\n %(t)s' %{'t':title}
#Print the result message
print (msg)
if plot:
self._PlotRegr(self.y, predict, title, color='maroon')
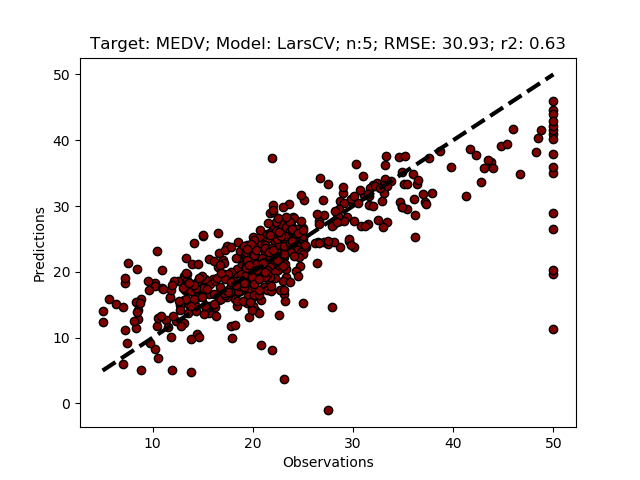
If you copy and paste the LarsCVFeatureSelectPipeline function to your module and run it, the results should be identical to the results of the LarsCVFeatureSelect. To run the function, call it from the __main__ section.
regmods.LarsCVFeatureSelectPipeline()
Embedding the pipeline in a cross validation search
In the two previous sections the threshold for selecting or discarding features from SelectFromModel was hardcoded to “mean”. But what if another threshold would give better results? You could test that by setting different thresholds, or create a loop that tested different thresholds. But that would take long time, and be tedious to write. The best option is to link different options to the pipeline, and test different options using GridSearchCV. To achieve that, you first create the pipeline and the different parameters you want to test, then wrap it together using GridSearchCV. The parameters that you want to test must be named using a strict convention: the first part must equal the name you set to the step (‘select’ and ‘regr’ in the codes above), followed by double underscores “__” and then the exact name of the hyper-parameter as defined for the process composing the step. When rapping the pipeline in a GridSearchCV you can still retrieve all the parameters from the included steps, as well as the results of the grid search. How to do that is explained and illustrated in the LarsCVFeatureSelectPipelineCV function.
def LarsCVFeatureSelectPipelineCV(self, plot=False):
'''LarsCV feature selection and regression using the full dataset
'''
#Set the model to LarsCV
mod = LarsCV()
name = 'LarsCV'
#Define SelectFromModel using the model (LarsCV) and a threshold
select = SelectFromModel(mod)
#Setup a pipeline linking the feature selection and the model
#The names will be used for linking parameters to each step
pipeline = Pipeline([
('select',select),
('regr',mod)
])
#Define the parameters to test in a grid search for optimizing hyper-parameters
#The first part of the key must correpspond to a step in the pipleine,
#and the last part must correspond to parameter accepted by the function defined in that step
parameters = dict(select__threshold=["0.5*mean","0.75*mean", "mean","1.5*mean","2*mean"])
#Define the grid search, set the pipeline as the estimator,
#and the parameters defined above as the param_grid
cv = GridSearchCV(pipeline, param_grid=parameters, verbose=1)
#Fit the cross validation search
cv.fit(self.X, self.y)
#All the parameters from each step in the bot the crossvalidation and the pipeline can be retrieved
#Retrieve and print the best results from the grid search
for i in range(1, 5):
candidates = np.flatnonzero(cv.cv_results_['rank_test_score'] == i)
for candidate in candidates:
print("Model with rank: {0}".format(i))
print("Mean validation score: {0:.3f} (std: {1:.3f})".format(
cv.cv_results_['mean_test_score'][candidate],
cv.cv_results_['std_test_score'][candidate]))
print("Parameters: {0}".format(cv.cv_results_['params'][candidate]))
print("")
#print the best resutls from the cross validation search
print 'cv best estimator:', cv.best_estimator_
print 'cv best score:', cv.best_score_
print 'cv best params:', cv.best_params_
print 'cv best index:', cv.best_index_
#Use the best estimator (cv.best_estimator) for retrieving the corresponding parameters in the pipleine
#Print the support (list of selected and discarded) features from the 'select' step (step = 0)
print 'step 0 support', cv.best_estimator_.steps[-2][1].get_support()
#Each step can also be accessed using its name
print 'step select support', cv.best_estimator_.named_steps['select'].get_support()
#Create a dictionary from the original X columns and the boolean support list identifying the selected features
selectD = dict(zip(self.columnsX, cv.best_estimator_.named_steps['select'].get_support() ))
#Extract the names of the selected features to a list
featureL = [item for item in selectD if selectD[item]]
#Print the selected features
print 'Selected features', featureL
#Get the number of features that was selected
nFeat = len(featureL)
#Print the parameters of the fitted regression
print 'fitted params', cv.best_estimator_.steps[-1][1].get_params()
#Print the coefficiens of the fitted regression
print 'coefficients', cv.best_estimator_.named_steps['regr'].coef_
#Predict the target
predict = cv.predict(self.X)
#Prepare a message for printing out the results
title = ('Target: %(tar)s; Model: %(mod)s; n:%(n)s; RMSE: %(rmse)1.2f; r2: %(r2)1.2f' \
% {'tar':self.target,'mod':name,'n': nFeat, 'rmse':mean_squared_error(self.y, predict),'r2': r2_score(self.y, predict)} )
msg = ' Regression results\n %(t)s' %{'t':title}
#Print the result message
print (msg)
if plot:
self._PlotRegr(self.y, predict, title, color='maroon')
Test the module by calling it from the __main__ section.
regmods.LarsCVFeatureSelectPipelineCV(True)
A more general function
The functions that you built so far in this module were written for understanding how a pipeline works, and how to embed a pipeline in a cross validation search. We can now construct a more general function that allows you to send any regressor through the pipeline and the cross validation.
def PipelineCVRegressors(self, plot=False, verbose=0):
'''Feature selection and regression with linear regressors with inbuilt CV
'''
for m in self.modelD:
print 'Model',m
#Define SelectFromModel using the model
select = SelectFromModel(self.modelD[m]['mod'])
#Setup a pipeline linking the feature selection and the model
pipeline = Pipeline([
('select',select),
('regr',self.modelD[m]['mod'])
])
#Define the parameters to test in a grid search for optimizing hyper-parameters
parameters = dict(select__threshold=["0.5*mean","0.75*mean", "mean","1.5*mean","2*mean"])
'''
if self.modelD[m]['param_grid']:
for p in self.modelD[m]['param_grid']:
param = 'regr__%(p)s' %{'p':p}
values = self.modelD[m]['param_grid'][p]
parameters[param] = values
print parameters
'''
#Define the grid search, set the pipeline as the estimator
cv = GridSearchCV(pipeline, param_grid=parameters, verbose=verbose)
#Fit the cross validation search
cv.fit(self.X, self.y)
#Retrieve and print the best results from the grid search
if verbose:
self.ReportSearch(cv,3)
#Create a dictionary from the original X columns and the boolean support list identifying the selected features
selectD = dict(zip(self.columnsX, cv.best_estimator_.named_steps['select'].get_support() ))
#Extract the names of the selected features to a list
featureL = [item for item in selectD if selectD[item]]
#Print the selected features
print ' Selected features', featureL
#Get the number of features that was selected
nFeat = len(featureL)
#Print the coefficiens of the fitted regression
print ' coefficients', cv.best_estimator_.named_steps['regr'].coef_
#Predict the target
predict = cv.predict(self.X)
#Prepare a message for printing out the results
title = ('Target: %(tar)s; Model: %(mod)s; n:%(n)s; RMSE: %(rmse)1.2f; r2: %(r2)1.2f' \
% {'tar':self.target,'mod':m,'n': nFeat, 'rmse':mean_squared_error(self.y, predict),'r2': r2_score(self.y, predict)} )
msg = ' Regression results\n %(t)s' %{'t':title}
#Print the result message
print (msg)
if plot:
self._PlotRegr(self.y, predict, title, color='maroon')
The results for each model tested thorugh the pipeline/cross validation is reported in a suppurt function, ReportCVSearch, that you must also add.
def ReportCVSearch(self, cv, n_top=3):
results = cv.cv_results_
for i in range(1, n_top + 1):
candidates = np.flatnonzero(results['rank_test_score'] == i)
for candidate in candidates:
print(" Model with rank: {0}".format(i))
print(" Mean validation score: {0:.3f} (std: {1:.3f})".format(
results['mean_test_score'][candidate],
results['std_test_score'][candidate]))
print(" Parameters: {0}".format(results['params'][candidate]))
print ' cv best estimator:', cv.best_estimator_
print ' cv best score:', cv.best_score_
print ' cv best params:', cv.best_params_
#Create a dictionary from the original X columns and the boolean support list identifying the selected features
selectD = dict(zip(self.columnsX, cv.best_estimator_.named_steps['select'].get_support() ))
#Extract the names of the selected features to a list
featureL = [item for item in selectD if selectD[item]]
#Print the selected features
print ' Selected features', featureL
#For models that expose "coef_", print "coef_"
if hasattr(cv.best_estimator_.named_steps['regr'],'coef_'):
#Print the coefficiens of the fitted regression
print ' coefficients', cv.best_estimator_.named_steps['regr'].coef_
Whether or not to print out the results for each model is determined by the parameter verbose. By default the results are not printed (verbose = 0).
The function PipelineCVRegressors returns the overall best model results.
To run the module using the PipelineCVRegressors function, you must define the regressors to use in the same manner that you did it in the previous posts. All the required functions are defined in the skeletion, and you just need to add and invoke the models in the __main__ section. The models I added below all have a built in cross validation for selecting features using cross validation. For three of the models I also added alternative settings for the hyper-parameter alphas. And for ‘ElasticNetCV’ I added a second set of options, for the parameter setting of l1_ratio .These alternatives will be evaluated by GridSearchCV. In the example below I also set plot to False, and verbose to 1. The best results, for each regressor, from the GridSearchCV will thus be printed, but not plotted.
modD = {}
modD['LarsCV'] = {}
modD['RidgeCV'] = {'alphas':[0.1, 1.0, 10.0]}
modD['LassoCV'] = {'alphas':[0.1, 1.0, 10.0]}
modD['LassoLarsCV'] = {}
modD['ElasticNetCV'] = {'alphas':[0.1, 1.0, 10.0], 'l1_ratio':[0.01, 0.25, 0.5, 0.75, 0.99]}
regmods.ModelSelectSet(modD)
bestModel,bestCV = regmods.PipelineLinearCVRegressors(0.3, False, 1)
print 'Best model:', bestModel
regmods.ReportCVSearch(bestCV, 1)
You can add any other model that you want to test, by importing it, including it in the model library in ModelSelectSet, and adding it to the modD dictionary in the __main__ section. If you want to use a cross validation search for tuning any hyper-paramter, add the hyper-parameter to the dictionary belonging to the model. As done for the hyper-parameter alphas above.
The complete code of the module that you created in this post is available at GitHub.
Resources
Pipeline as Sci-kit learn.
Workflows in Python by Katie M.
Scikit learn regressors with built in cross validaton.
Completed python module on GitHub.